Gene Editing Primer: The 'How', the 'If' and the 'When'
July 16, 2015
by Jennifer Phillips, PhD
Over the past few decades, our ability to mine the wealth of information encoded in the human genome—in addition to the genomes of many other species—has increased exponentially. We now understand a staggering amount about how information is stored in the DNA code, how the code provides the blueprint for building molecules that perform needed functions within the body, and how changes to parts of the code can manifest as human diseases. This fundamental understanding of the information written in our genes is a prerequisite to developing ways to intervene when genetic problems are identified, and we are now at the leading edge of using this information to devise progressively more targeted therapies for inherited disorders. A few strategies, including the virally delivered gene replacement therapy for USH1B that’s currently in clinical trials, are already underway, but a new potential therapy method using something called CRISPR/Cas gene editing is getting increasing coverage in the press, so it seems like a timely topic for a blog post here.
What is “CRISPR/Cas” and why is everyone so excited about it?
Simply put, this is a tool that combines targeting information that homes in on a precise location within the genome (that’s the CRISPR part), with the ability to generate DNA breaks at that site once it arrives there (that’s Cas’s job). Such tools are integral to the process of gene editing, because the ability to interrupt a continuous strand of DNA at a specific location is the first step to altering the information encoded there. There are other gene editing tools currently in use in research labs, but CRISPR/Cas has quickly gained popularity for the very practical reason that it’s a bit easier and less expensive to use than the other options.
Using the CRISPR/Cas system as a tool is a relatively new phenomenon, but the system itself is ancient. Years ago, researchers studying bacteria noticed that many bacteria had mysterious regions of repeated short DNA sequences in certain parts of their genomes. These repetitive sequences appeared in clusters and were actually palindromes, constructed of a sequence that read the same forwards and backwards. Additionally, each repetitive sequence in the cluster was separated by a short sequence of DNA that didn’t seem to be repeated elsewhere. The name ‘CRISPR’ is derived from all these observed characteristics: Clustered Regularly Interspaced Palindromic Repeats. This mysterious pattern intrigued a number of scientists, so it became a relatively well-studied phenomenon. A subsequent observation identified that these CRISPR repeats were always in close proximity to genes that belonged to the same class, which became known as ‘Cas’ ‘CRISPR-associated genes. All Cas genes encode variations of a type of protein, an enzyme, that can cut DNA strands. The final piece of the puzzle was the realization that the unique short DNA sequences separating the CRISPR repeats in bacterial genomes were highly similar to DNA sequences of viruses.
With this collection of knowledge, researchers came up with the extraordinary explanation for the function of CRISPRs: they are a form of immunity against viruses. Viruses, as you know, propagate themselves by entering a host cell and then co-opting the native DNA replication machinery to make more copies of themselves. It turns out that when a virus invades a bacterial cell, the cell copies a little of the viral DNA and stores it in its own genome, flanked by CRISPRs to create a genetic memory of the encounter. This cached viral code can then be used to identify and destroy a similar viral invader down the road, by producing a small molecule that can recognize and bind to the viral sequence and can also physically attach to a Cas enzyme. The ‘guide’ molecule then travels through the cell hunting for the viral sequence it’s built to recognize and bringing along the means to destroy it.
When the ‘guide’ part of the CRISPR recognizes the viral DNA sequence it’s coded to find, it locks on. Once in position, the associated Cas enzyme cuts right through the viral DNA sequence to prevent it from replicating, and the pathogen is neutralized. Cool, huh?
As soon as researchers worked out the details of how this system functions in bacteria, they could immediately see that it would be a useful tool for genetic research everywhere, and the ensuing years since the mystery of CRISPR/Cas was unlocked have been spent trying to harness its editing powers for good.
So, what exactly can researchers do with a tool that induces breaks at a specific location on the DNA strand? Well, the list of possibilities is growing longer even as we speak, largely due to the versatility this system offers. In the original CRISPR/Cas construction, guide sequence recognizing a viral gene is hooked up to the DNA cutting protein. But the guide sequence doesn’t have to correspond to a viral gene sequence, it can be made to target virtually any DNA sequence. Researchers can make a guide molecule that will recognize a chosen region of a chromosome, while also giving it the properties necessary to associate with a Cas protein. If the guide sequence finds its way to the target, the Cas enzyme will do its work and cleave the DNA strand right there.
Once cuts in the DNA strand initiate an innate repair mechanism that reconnects the open ends of the molecule in a way that sometimes includes or excludes random bits of information. The fancy science term for this process is non-homologous end-joining (NHEJ).
Imagine cutting through a string of beads, and then trying to repair it as quickly as possible. In some cases, you might be able to rejoin the two broken ends without losing any beads. Whew! No harm done.
In other cases, though, it might be necessary to re-string some beads. Here’s the problem, though, you don’t remember exactly which beads went where, or even how many there were on the original strand. If you don’t manage to get them all restrung, the repaired necklace will be a bit shorter. Or, you might find extra beads in addition to the ones that dropped off your necklace, and accidentally include them in the strand, making it slightly longer as a result. In either case, the repaired necklace will be different after the fact, due to your random re-stringing efforts.
This is more or less what happens when genes are mutated by natural causes. Information is lost, or changed, and the final product doesn’t work properly. The ability to create targeted mutations in the laboratory is an incredible research tool for understanding how things go wrong in genetic diseases. By inducing breaks in the DNA at specific points in a given gene, we can now study how mutations in different genes, or different parts of a single gene, affect the protein coding information and contribute to the cellular or molecular dysfunction that leads to disease.
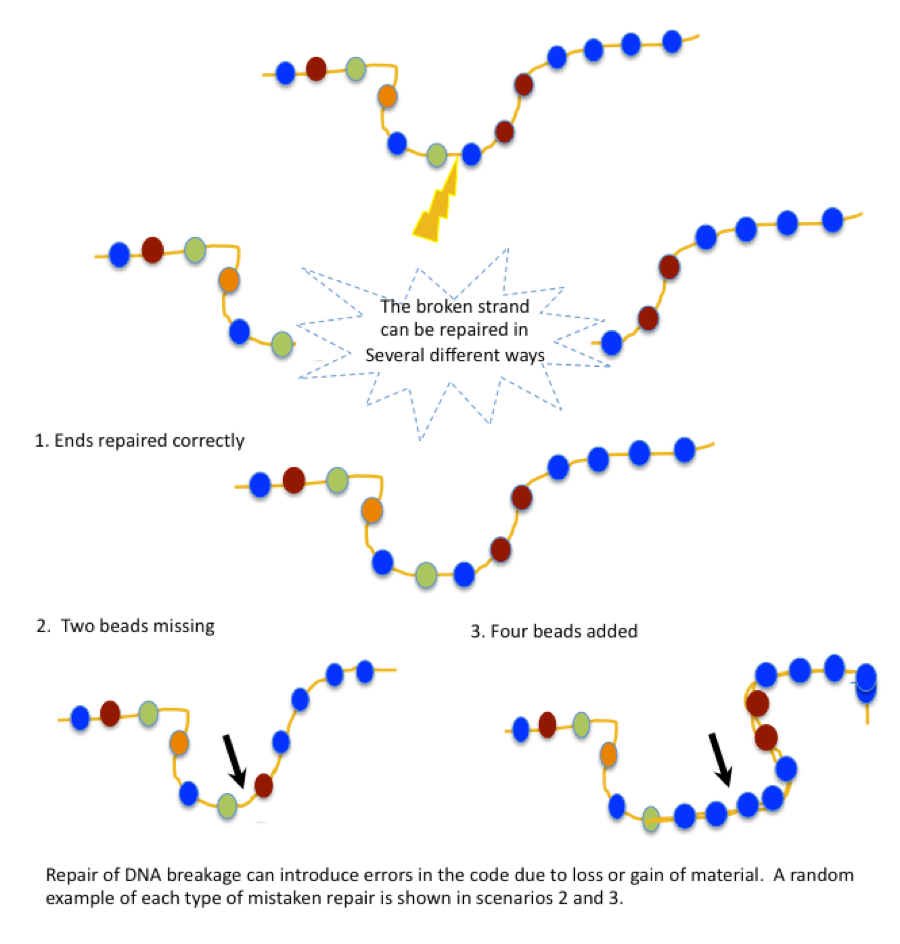
Image Credit: Dr. Jennifer Phillips
In addition to this unprecedented ability to target and study disease genes, CRISPR/Cas has promising potential for disease therapy as well. Some diseases result when the abnormal proteins created based on genetic mutations interfere with normal cellular function in some way. In these cases, the CRISPR/Cas system as described above could potentially be used to disable the gene so that this disruptive protein would no longer be made.
An even greater therapy potential, however, comes from additional gene editing methods that are possible when the continuity of the DNA strand is disrupted at a specific location. The main feature of simply cutting the DNA and allowing NHEJ to commence is that random mistakes will inevitably occur and a loss of gene function will result.
But, consider the bead restringing analogy again. What if, instead of leaving it to chance, someone provided you with an example of how the beads should be strung. You observe that some of the bead pattern in the example exactly matches the pattern on the pieces of your broken necklace, and this convinces you that the rest of the pattern must be a good match as well. You don’t remember the exact pattern of the original necklace, so you’re going to trust the source and reconstruct the necklace based on the sequence provided. In this case, though, the helpful person has actually provided you with a pattern that’s different from the original. When the necklace is restrung, the sequence of the beads is different.
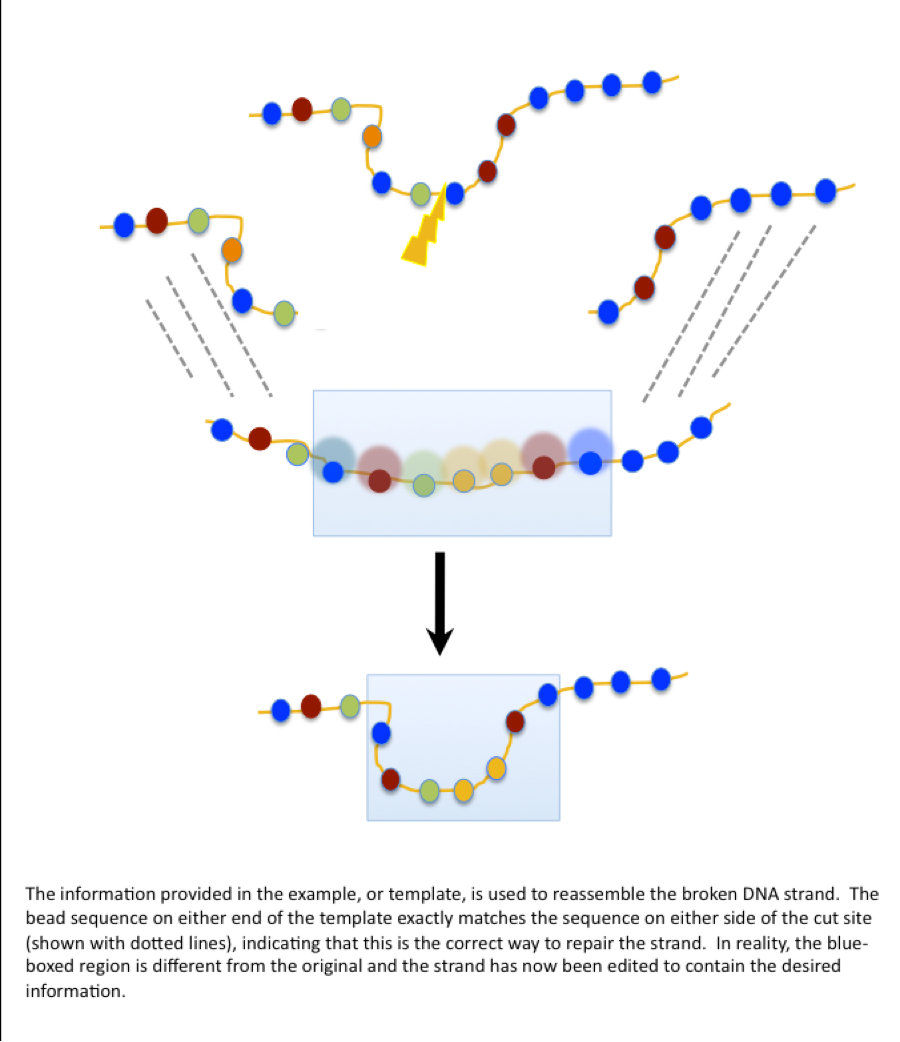
Image Credit: Dr. Jennifer Phillips
When applied to DNA, this is referred to as homology-directed repair (HDR) and is the essence of what we’re talking about when we refer to “gene editing”. The original code is replaced with new information based on a template that matches up with the intact parts of the strand on either side of the break, but contains an alternate sequence in the middle. The potential clinical application of this method, as you may have guessed, is in replacing the faulty code of disease genes with correct information. The CRISPR/Cas system could be used to break the DNA strand at the site of a genetic mutation. A template sequence containing the corrected information could be provided at the same time, and HDR of the new strand could edit out the original disease-causing mutation.
This is an exciting possibility, no question about it, but do note the number of times I’ve used words like ‘potential’ and ‘may’ and ‘could’. There are numerous logistical issues to solve, not to mention ethical issues to contemplate, before this type of gene editing would be suitable to try in people.
Let’s tackle the logistics first. As you have (hopefully) gathered from the ‘string of beads’ illustrations above, there is quite a bit of random chance involved in how things play out with this molecular repair mechanism. CRISPR/Cas systems have been used extensively for the past several years on many different model organisms, as well as human tissue cultures, so collectively we have a good idea of how it works and what the variables are, so it’s a common expectation at this point in the development of CRISPR/Cas methods that there will be quite a bit of variation with each attempt. A separate copy of the genetic code of each individual organism-- all our chromosomal DNA—is housed in the nuclei of nearly every cell of our bodies. When CRISPR/Cas9 materials are introduced into a multicellular system, therefore, the editing occurs independently in every cell. This means that, not only are the effects different from organism to organism, but are also different within a single organism.
In the lab, we’re able to take DNA samples from the organisms we’ve ‘CRISPRed’, and analyze them to see what kind of changes occurred at our target site. These results allow us to identify the types of DNA changes that we meant to make, but also reveal a considerable number of novel mistakes. We see deletions or insertions of information at the desired location that change the code in undesirable ways. We see the integration of new information based on the provided template that overwrites the original code, but in some cases there’s an additional bit of new code included that disrupts the information and therefore won’t restore the correct genetic information as intended.
Additionally, there are some times that the CRISPR/Cas system finds the wrong target. The sequence recognized by the CRISPR guide is very short, relative to the vast amount of DNA found in every cell. The guide sequence is usually unique, but occasionally, there is sequence somewhere else in the genome that might be similar enough to the target that the CRISPR will find that, as well, and make a change to a completely different region by accident.
At the level of basic and preclinical research, screening techniques allow us to select only the experiments that worked, and chuck the rest. But the same type of screening wouldn’t be possible if the technique were applied directly to the cells of a living person. Anything designed for use in living humans needs to be far more reliable and controllable that the current system.
That said, although we can’t screen the cells of living people when they’re IN people, we can screen cells in culture that have been treated with CRISPR/Cas. This technique has the greatest potential to pay dividends for human disease treatment in the shortest period of time. I’ve written before about the work on patient-derived induced pluripotent stem cells (iPSCs) that enable scientists to culture cells obtained from individual patients and coax them to differentiate into complex tissues, like retina. Beyond the unique opportunity to learn more about the molecules causing particular variations of a given disease, there is some hope that these cultured, patient-derived cell populations will someday be used for cell replacement-based therapies. In that context, cells obtained from a patient could be targeted for CRISPR/Cas gene editing in culture. Subsequently, the treated, cultured cells could be screened for the corrected version of the gene and for unintended ‘off-target’ effects, and the cells whose DNA shows only the right editorial changes could then be prepared for implantation back into the patient. Again, this is still years away, but the potential is there and efforts to optimize this method are proceeding as we speak.
That brings us to the ethical ramifications of modifying the human genome. The above scenario describes a situation where changes are being made to tissue taken, with consent, from individual patients. But most CRISPR/Cas work to date has involved making changes to the embryonic genomes of model organisms, like mice and zebrafish. It‘s always been a possibility, therefore, that the treatment could be applied to human embryos in the same manner, and recently, a paper was published by a research group in China that did just that: CRISPR/Cas treatment of human embryonic stem cells.
The human ES cell line used was chosen specifically because it wasn’t viable. It carried an abnormal number of chromosomes, and therefore would not have been able to develop into a human fetus, even under optimal conditions. Still, the ethical issues loomed so large in this work that several prestigious journals rejected the work for fear of endorsing a research direction that many aren’t comfortable with.
Ethical issues aside, though, the results of the experiment were fairly unremarkable, in that they recapitulated all the phenomena we’re already familiar with in the animal models. Editing mistakes were made, both at the target site and in off-target regions, and the DNA was edited differently from one cell to another. The authors, unsurprisingly, concluded that this technique is just as imperfect in human cells as it is in every other cell type tested thus far, and if anything solidified the consensus that we still have a lot to learn before this technique is ready for use in humans.
While it will clearly take considerable time and effort to optimize this system and establish ethical guidelines for therapeutic use, the potential applications for treating genetic diseases using CRISPR/Cas are phenomenal. There is reason to hope that this strange bacterial innovation could someday provide a cure for human diseases like Usher syndrome.
For further reading on the discovery and applications of CRISPR/Cas, here are links to some great articles on the topic:
http://gizmodo.com/everything-you-need-to-know-about-crispr-the-new-tool-1702114381
https://www.quantamagazine.org/20150206-crispr-dna-editor-bacteria/
http://phenomena.nationalgeographic.com/2015/04/22/editing-human-embryos-so-this-happened/







